Q: Are there any samples that should be excluded on the basis of poor quality or too few counts?
A: No. The reports we should be concerned with (Per Base Sequence Quality, Per Base N Content, Sequence Length Distribution and Adapter Content) receive passing (Green) scores for all samples, both forward and reverse reads.
Q: Do Illumina adapters need to be removed?
A: No, as above, passing scores for all files.
Q: What are the recommended truncation parameters to use?
A: Forward truncation at 89, reverse truncation at 186.
Q: What is the expected retention percent for this filtration step?
A: 95.24 %.
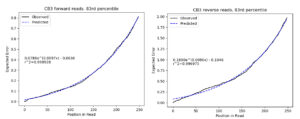
Q: Regarding the FIGARO plots, are predicted error rates good or bad?
A: Very good, r-squared values over 99%.
Q: From viewing the table in denoising-stats.qzv, what is the mean overall retention?
A: 74.488 %. This is for the percentage of input reads passing all steps: filtering, denoising, merging and chimera removal. The above value of 95.24 % is for reads passing the filtration step only.
Q: From viewing the quality plots in demux.qzv, would you have chosen different truncation parameters? If so, did they give better overall retention?
A: I cannot answer this for you. The question is meant for you to explore the results of alternative choices. If you did better than FIGARO, please let me know (see the Contacts page).
Q: How does the merging retention in denoising-stats.qzv compare with FIGARO’s prediction?
A: FIGARO’s retention per cent is for reads passing the filters, not for merging. FIGARO’s prediction of 94.25 is very close to the observed value of 95.251 (mean for all samples).
