Compact letter displays (CLDs) are letters that show which treatment groups are not significantly different by some statistical test. It is often desirable to include CLDs on graphs. Here I show how to add them to a box plot created with ggplot2.
First, make an example plot using the iris data:
library(ggplot2)
library(car)
library(QsRutils)
data("iris")
plt1 <- ggplot(data = iris, aes(x=Species, y=Petal.Length)) + geom_boxplot()
plt1

I want to add CLDs just above the tops of the box whiskers in the plot. I can use the base boxplot function to capture the coordinates for these points.
box.rslt <- with(iris, graphics::boxplot(Petal.Length ~ Species, plot = FALSE)) str(box.rslt) List of 6 $ stats: num [1:5, 1:3] 1.1 1.4 1.5 1.6 1.9 3.3 4 4.35 4.6 5.1 ... $ n : num [1:3] 50 50 50 $ conf : num [1:2, 1:3] 1.46 1.54 4.22 4.48 5.37 ... $ out : num [1:2] 1 3 $ group: num [1:2] 1 2 $ names: chr [1:3] "setosa" "versicolor" "virginica"
The fifth row of box.rslt$stats gives the y coordinates for the tops of the whiskers.
box.rslt$stats [,1] [,2] [,3] [1,] 1.1 3.30 4.50 [2,] 1.4 4.00 5.10 [3,] 1.5 4.35 5.55 [4,] 1.6 4.60 5.90 [5,] 1.9 5.10 6.90
Next I have to get a vector of the letters to add to the plot. For this example, I will make a pairwise t-test which outputs a matrix of p-values.
ptt.rslt <- with(iris, pairwise.t.test(Petal.Length, Species, pool.sd = FALSE))
Looking at the structure of ptt.rslt, we see that ptt.rslt$p.value gives a matrix of p.values:
str(ptt.rslt)
List of 4
$ method : chr "t tests with non-pooled SD"
$ data.name : chr "Petal.Length and Species"
$ p.value : num [1:2, 1:2] 1.99e-45 2.78e-49 NA 4.90e-22
..- attr(*, "dimnames")=List of 2
.. ..$ : chr [1:2] "versicolor" "virginica"
.. ..$ : chr [1:2] "setosa" "versicolor"
$ p.adjust.method: chr "holm"
- attr(*, "class")= chr "pairwise.htest"
ptt.rslt$p.value
setosa versicolor
versicolor 1.986887e-45 NA
virginica 2.780888e-49 4.900288e-22
From this matrix we can use QsRutils::make_letter_assignments to get a vector of letters for our CLDs.
ltrs <- make_letter_assignments(ptt.rslt)
str(ltrs)
List of 3
$ Letters : Named chr [1:3] "a" "b" "c"
..- attr(*, "names")= chr [1:3] "setosa" "versicolor" "virginica"
$ monospacedLetters: Named chr [1:3] "a " " b " " c"
..- attr(*, "names")= chr [1:3] "setosa" "versicolor" "virginica"
$ LetterMatrix : logi [1:3, 1:3] TRUE FALSE FALSE FALSE TRUE FALSE ...
..- attr(*, "dimnames")=List of 2
.. ..$ : chr [1:3] "setosa" "versicolor" "virginica"
.. ..$ : chr [1:3] "a" "b" "c"
- attr(*, "class")= chr "multcompLetters"
ltrs$Letters
setosa versicolor virginica
"a" "b" "c"
ltrs$Letters gives the vector of letters that we want. Now we can make a data frame to add the CLDs to the box plot.
x <- c(1:length(ltrs$Letters))
y <- box.rslt$stats[5, ]
cbd <- ltrs$Letters
ltr_df <- data.frame(x, y, cbd)
ltr_df
x y cbd
setosa 1 1.9 a
versicolor 2 5.1 b
virginica 3 6.9 c
If we plot the CLDs at the coordinates in ltr_df, they will over plot the tops of the whiskers . We need to nudge the CLDs upward to avoid the overlap. To determine how much to nudge, I will get the range of the Y-axis and nudge upward 5% of this range.
lmts <- get_plot_limits(plt1) y.range <- lmts$ymax - lmts$ymin y.nudge <- 0.05 * y.range plt1 + geom_text(data = ltr_df, aes(x=x, y=y, label=cbd), nudge_y = y.nudge)
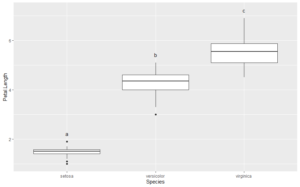
The CLDs are perfectly positioned, and without any trial and error.
